Institut für Genetik
Im Mittelpunkt der Forschung am Institut für Genetik stehen dynamische Prozesse, wie Zellmigration, Zellteilung und Signalverarbeitung. Zellmigration und Zellteilung sind elementare Parameter bei Entwicklungsprozessen, Regeneration und Homöostase. Signalverarbeitung zwischen Nervenzellen und die Modellierung dieser Prozesse gibt uns Einblicke in die Funktion komplexer neuronaler Netzwerke.
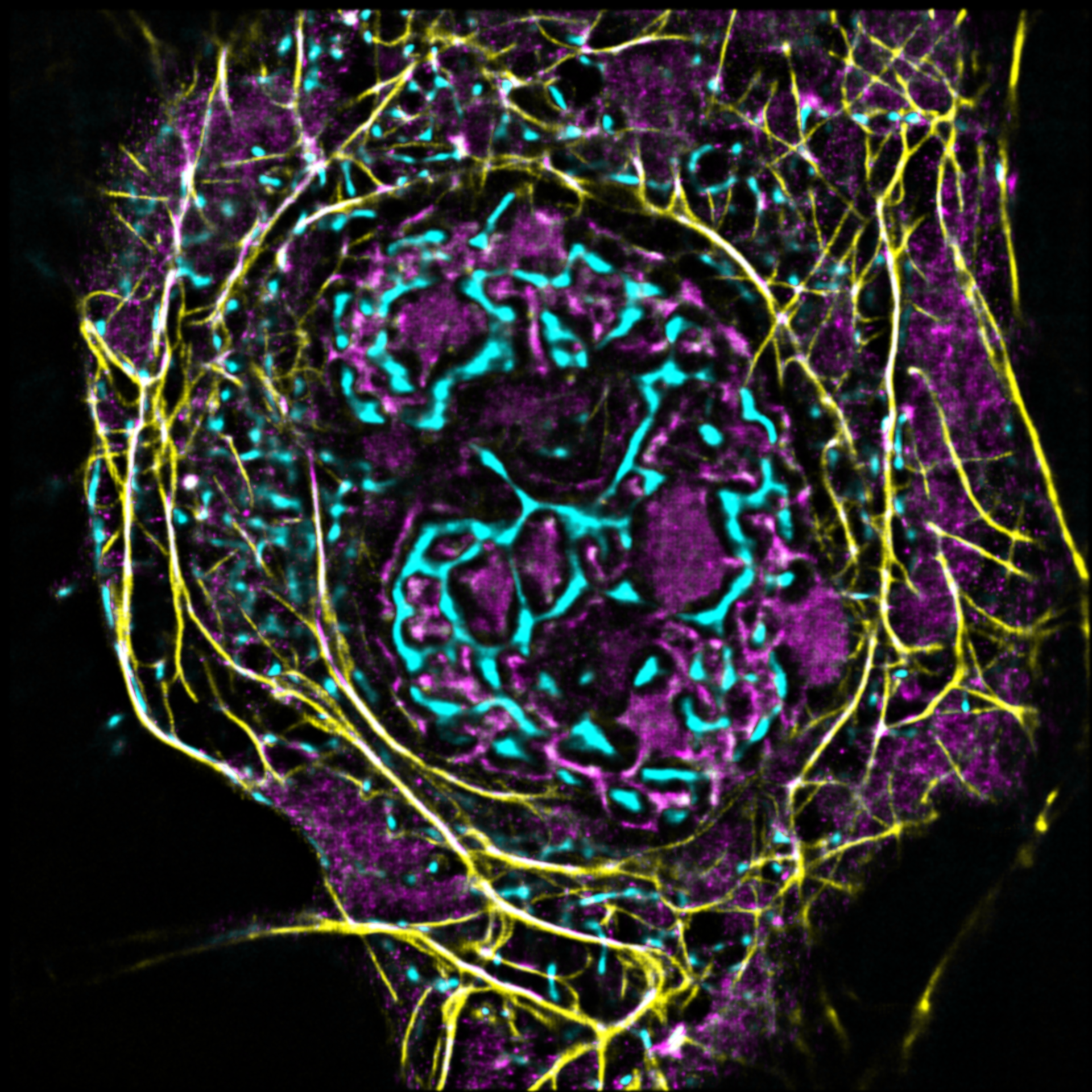
AG Witke:
Zellmigration
Unsere Arbeitsgruppe untersucht die molekularen Grundlagen von Zellmotilität auf den Ebenen des Zytoskeletts.
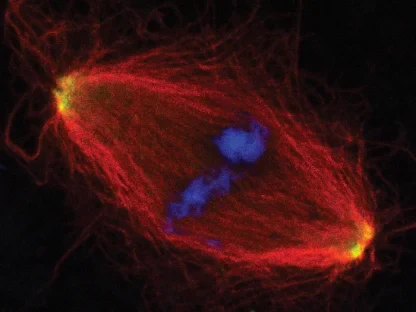
AG Gruß:
Zellteilung
Die AG Gruß untersucht die Funktion von Mikrotubuli in der Zellteilung und der Assemblierung von RNPs in der Interphase.
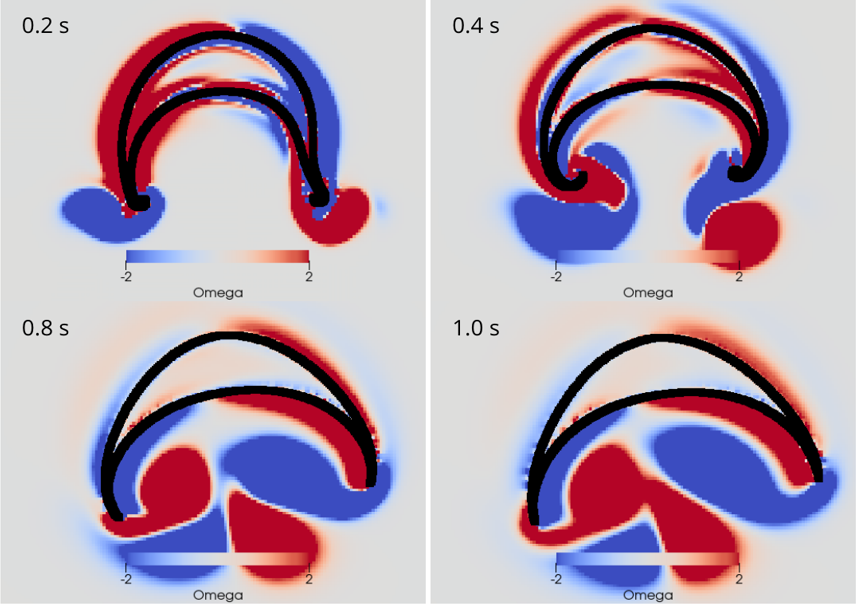
AG Memmesheimer:
Computergestützte Biologie
Die AG Memmesheimer untersucht die Dynamik und Informationsverarbeitung von biologischen neuronalen Netzwerken mit Hilfe von Modellierungen.
Mögliche Themen für Abschlussarbeiten oder Forschungspraktika finden Sie auf den Seiten der beiden Arbeitsgruppen.

Abschlussarbeiten & Promotion
Wir sind stets auf der Suche nach motivierten Mitarbeitern. Wenn Sie Interesse haben, an einem unserer spannenden Projekte mitzuarbeiten, schreiben Sie uns an.
Kontakt
Geschäftszimmer: Simone Christian
AG-Leiter: Prof. Dr. Oliver Gruss, Prof. Dr. Walter Witke, Prof. Dr. Raoul-Martin Memmesheimer
Koordination Lehre: Dr. Michael Reinke
IT Service: Stefan Klein
Das Institut für Humangenetik am Universitätsklinikum Bonn finden Sie hier:
https://www.humangenetics.uni-bonn.de/10
Adresse
Raum 3.005
Karlrobert-Kreiten-Straße 13
53115 Bonn





Links
- https://www.biologie.uni-bonn.de/genetik/de/genetik#Forschung
- https://www.biologie.uni-bonn.de/genetik/de/genetik#Lehre
- https://www.biologie.uni-bonn.de/genetik/de/genetik#Kontakt
- https://www.biologie.uni-bonn.de/genetik/personenverzeichnis-der-genetik
- https://www.biologie.uni-bonn.de/genetik/de/arbeitsgruppen/ag-witke
- https://www.biologie.uni-bonn.de/genetik/de/arbeitsgruppen/ag-gruss
- https://www.biologie.uni-bonn.de/genetik/de/arbeitsgruppen/ag-memmesheimer
- https://www.biologie.uni-bonn.de/genetik/de/lehre/lehre_bachelor
- https://www.biologie.uni-bonn.de/genetik/de/lehre/lehre_master
- https://www.humangenetics.uni-bonn.de/
- https://www.biologie.uni-bonn.de/genetik/de#
- https://www.openstreetmap.org/copyright